This is the latest blog in our series on the PatCID Chemical Structure Database, a tool we’re testing in collaboration with IBM Research’s team. We’ll be hosting a webinar in the early fourth quarter of this year with Dr. Gerhard Ingmar Meijer, one of the tool developers, who will provide an introduction and live demonstration of PatCID.
In a rush? Here’s the summary:
Markush structures are often used in patents. They define a fixed molecular core and list various possible chemical groups that can be attached, allowing a single patent to protect many related compounds. This approach is often useful in intellectual property but makes it challenging and expensive to search for these structures accurately. To solve this, IBM Research’s team in Zurich has developed a new AI model, MarkushGrapher. This model will soon become a feature of their PatCID tool. It uses a combination of image and text analysis to interpret Markush structures, which IBM expects will provide for a faster and more scalable alternative to traditional manual indexing methods.
What is PatCID?
PatCID (Patent-extracted Chemical-structure Images database for Discovery) is an automated chemical structure indexing tool and database developed by IBM’s Deep Search team. The team’s goal is to identify and annotate chemical structures in patent publications more accurately than existing automated tools, while offering a faster, more cost-effective alternative to the manually annotated databases that remain the industry standard. PatCID’s first release, covering specific chemical structures, became available last year. TPR has been working with IBM’s team to assess the tool, and you can read our earlier articles for additional context here. Early next year, IBM plans to add Markush structure indexing and search capabilities, one of the most challenging areas to automate. The rest of this article covers that upcoming update.
Here we provide a brief explanation of Markush structures in searching and their use in patents, or scroll further to find out how PatCID is approaching the challenge of automating the indexing of Markush structures using the latest AI techniques.
How Markush Structures Represent Variants
A Markush structure typically has two main parts:
- Core (Backbone) – the fixed part of the molecule, shown as a chemical drawing.
- Variable groups (R-groups or substituents) – placeholders for one or more possible chemical groups. These are usually described in the text of the patent or in a substituent table.
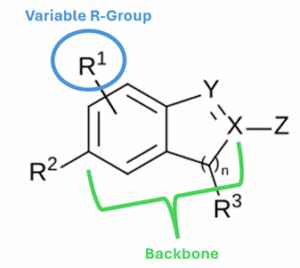
Image 1. Example Markush structure. The R-Groups would be defined elsewhere in the patent.
The total number of possible molecules is the product of all possible combinations of the variations at each position. For example, if there are three R-groups (like in the image above) and each can be one of three different substituent variants, then there are 27 (33) possible variants described. In this way, a small table of possibilities can quickly expand into hundreds or thousands of combinations.
Additional complexity comes from:
- Positional variation – substituents can appear at different positions on the backbone.
- Frequency variation – substituents can appear more than once.
- Combinatorial explosion – when multiple points of variation generate millions of possible structures.
Why Use Markush Structures in Patents
Markush claims let applicants protect an entire class of related compounds, not just a single molecule. This can:
- Stake out territory around a chemical scaffold that may show similar biological activity across variants.
- Cover potential drug candidates when it’s unclear which variant will go to market.
- Make it harder for competitors to design around the invention, as even unsynthesized variants within the defined scope may be protected.
- Reduce disclosure clarity for competitors, strategically limiting how easy it is to pinpoint the exact lead compound.
Implications for Searching
Markush structures are difficult to search because they’re split between:
- A graphical backbone, which requires chemical structure recognition.
- Textual substituent descriptions, which must be interpreted and matched to the backbone.
Manual indexing by chemistry experts is the gold standard for Markush structure indexing accuracy. However, the manual indexing process can be slow and costly, making these resources expensive to use and often creating a long lag before new publications are searchable.
Despite the challenges, Markush searching is essential for:
- Freedom to Operate (FTO): Determining whether a compound falls within an existing Markush claim.
- Invalidity: Finding earlier disclosures of similar compounds that could challenge a patent.
- Patentability: Establishing that another patent applicant has not previously described a compound within a Markush claim.
Automated Markush Indexing – The challenge
Automated indexing can process far more publications at lower cost than expert human chemists, but usually with lower accuracy. Markush structures add complexity because they are multimodal: as discussed above, the fixed chemical backbone is shown in a diagram, while the variations (R-groups) appear in text or tables. Previously, automated algorithmic interpretations of both input types have not been very successful in terms of accuracy.
An Additional Challenge: The Data Gap
PatCID’s approach relies on machine learning, which requires large, high-quality labeled datasets. For Markush structures, these datasets were scarce. To address this, the team created:
- M2S Benchmark Dataset – the first (openly available) manually annotated set of real Markush structures from USPTO, EPO, and WIPO patents.
- Synthetic Data Pipeline – generates realistic Markush examples from PubChem molecules, with variations modeled on real-world data.
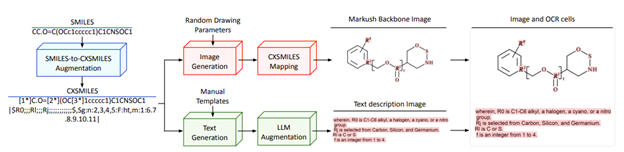
Figure 1. From Morin, et al., describes the Synthetic Data Pipeline used to generate training samples.
The MarkushGrapher model
Detailed in MarkushGrapher: Joint Visual and Textual Recognition of Markush Structures (Morin, et al., 2025), the system combines:
- Vision-Text-Layout (VTL) encoder – processes images, OCR text, and spatial layout.
- Optical Chemical Structure Recognition (OCSR) encoder – extracts the chemical backbone as a graph.
The two outputs are combined to generate a CXSMILES backbone (an extended line notation that encodes both molecules and structural annotations) along with substituent tables. Cross-modal reasoning lets the model recover missing details from one source using information from the other.
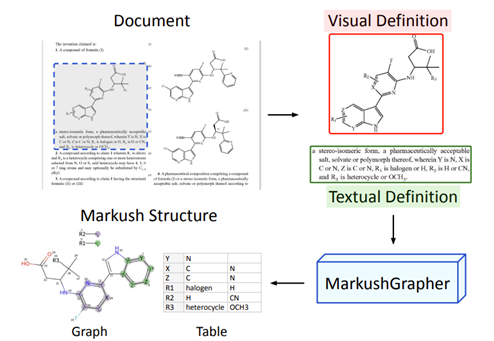
Figure 2. From Morin, et al., describing Markush structures being identified in patents, then the Visual and Textual definitions being encoded and combined by MarkushGrapher to produce the CXSMILES Graph and Substituent Table.
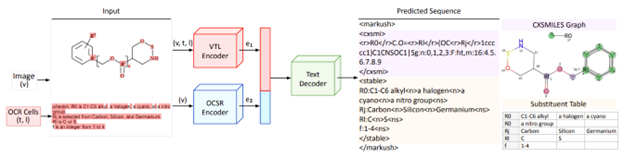
Figure 3. From Morin, et al., expanded description of the MarkushGrapher combining the input image and text from the VTL and OCSR Encoders to generate the CXSMILES Graph and Substituent Table.
Performance
MarkushGrapher was tested against image-only tools (DECIMER, MolScribe) and multimodal models (Pixtral-12B-2409, Llama-3.2-11B-Vision-Instruct, GPT-4o, Uni-SMART) on synthetic and real-world datasets. It outperformed all listed tools on nearly every metric and excelled in recognizing positional and frequency variations.
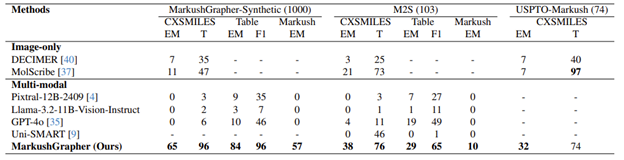
Table 1. From Morin, et al., shows the results of the comparison of MarkushGrapher with existing MMSR (Multimodal Markush Structure Recognition) models. EM = Exact Match, T = Tanimoto score, F1 = F1-score; similarity score between 0 (most dissimilar) and 100 (most similar).
Final Thoughts – TPR’s Review
In a nutshell, PatCID MarkushGrapher appears to have come a long way in providing annotation of Markush structures in patent publications; and a further improved version of MarkushGrapher will become available towards the end of the fourth quarter of this year. While compiling the earlier datasets of specific chemical structures, the IBM team already identified and saved hundreds of millions of Markush structures, which are now just waiting to be processed. If integrated into PatCID, the MarkushGrapher model could offer a faster, more scalable option for Markush searching, bridging the gap between costly manual indexing and less accurate automated tools.
To repeat TPR’s position from earlier articles, our ongoing assessment of PatCID suggests it’s a promising tool for chemical patent searching. While it does not replace established, industry-standard solutions, its automated approach could potentially offer a valuable, fast, and scalable option for initial screening and analysis. The upcoming integration of the MarkushGrapher function is poised to significantly enhance its capabilities, tackling one of the most complex challenges in patent searching. We will continue to evaluate PatCID’s performance and look forward to sharing more insights with you. Don’t forget to join our webinar in the early fourth quarter for a live introduction and demonstration of the tool.
Have questions regarding chemical patent searching and want to discuss with our team, please contact us.
Resources and Further Reading:
- Simmons, E.S. Markush structure searching over the years. World Patent Information. Elsevier, 2003.
- ACD/Labs. What are Markush structures, and how can I draw them? Visited August 12, 2025. https://www.acdlabs.com/blog/markush-structures-what-they-are-and-how-to-draw-them/
- Wikipedia. Markush structure. Last modified August 8, 2025. https://en.wikipedia.org/wiki/Markush_structure
- Markush, E.A.; Pharma Chemical Corp. Pyrazolone dye and process of making the same. US Patent 1,506,316, filed March 22, 1922, issued August 26, 1924. https://patents.google.com/patent/US1506316A/en
- Morin, T., et al. MarkushGrapher: Joint Visual and Textual Recognition of Markush Structures. arXiv preprint, visited March 20, 2025. https://arxiv.org/abs/2503.16096
© Technology & Patent Research International, Inc.


